16 510: Maintaining your Bioconductor package
16.1 Instructor name and contact information
- Nitesh Turaga40 (nitesh.turaga@roswellpark.org)
16.2 Workshop Description
Once an R package is accepted into Bioconductor, maintaining it is an active responsibility undertaken by the package developers and maintainers. In this short workshop, we will cover how to maintain a Bioconductor package using existing infrastructure. Bioconductor hosts a range of tools which maintainers can use such as daily build reports, landing page badges, RSS feeds, download stats, support site questions, and the bioc-devel mailing list. Some packages have their own continuous integration hook setup on their github pages which would be an additional tool maintainers can use to monitor their package. We will also highlight one particular git practice which need to be done while updating and maintaining your package on out git system.
16.2.1 Pre-requisites
Accepted Bioconductor package or plans to contribute a Bioconductor package in the future.
16.2.2 Participation
Students will be expected to follow along with their laptops if they choose to, although it is not needed.
16.2.3 Time outline: 50 mins short workshop
- Introduction: 10 mins
- Why maintain your package
- Infrastructure used for maintenance
- Outline how to use infrastructure: 35 mins
- Build report
- Version numbering
- Landing page badges
- Download statistics
- RSS feeds
- Support site, Bioc-devel mailing list
- Github
- Sync package with Github / Bioconductor before every change
- Github issues
- Round up of resources available: 5 mins
- Acknowledgements
16.2.4 Workshop goals and objectives
- Gain knowledge of how to track Bioconductor packages via download statistics, RSS feeds.
- Understand the importance of supporting their user community and how to do so using the support site and bioc-devel mailing list.
- Maintain their package and fix bugs using build reports, continuous integration tools.
- Update their package consistently on both Github and Bioconductor private git server.
16.3 Introduction
Maintainer - Who should people contact if they have a problem with this package? You must have someones name and email address here. You must make sure this is up to date and accurate.
Maintaining it is an active responsibility undertaken by the package developers and maintainers. Users contact developers for various reasons.
Keep your email up to date in the DESCRIPTION file!
Maintainer: Who to complain to <your_fault@fix.it>or
Authors@R: c(
person("Complain", "here", email="your_fault@fix.it", role=c("aut", "cre"),
person("Don't", "Complain", role="aut"))
)NOTE: There should be only 1 maintainer (one “cre”). But all authors get credit for the package.
Interchangeable terminology
Bioconductor maintainers <-> Bioconductor developers <-> Package developer <-> developer
16.3.1 Why maintain your package
Changes in underlying code (upstream dependencies of your package) might break your package.
Packages using web resources might fail because of change of location of these resources.
Bug fixes reported or discovered due to usage.
Feature requests from users.
Improvements in the science behind your package, which lead to updates.
It leads to wide spread usage of your package.
It’s required, also a good practice that users appreciate.
Poorly maintained packages get deprecated if they fail build and check on Bioconductor during release cycles. (Bioconductor is fairly serious about this)
16.3.2 Infrastructure used for maintenance
Build machines are dedicated towards building your package daily. As a maintainer it’s useful to check your package build report once a week, both in the current RELEASE and DEVEL cycle.
http://bioconductor.org/checkResults/
Build machines cover three platforms (Linux, Windows and OS X) over RELEASE and DEVEL versions of Bioconductor.
Version control - Since last year, GIT has been the primary version control system.
16.4 Outline how to use infrastructure
Everything you need is on the Bioconductor website. As a developer the link with most important resources,
http://bioconductor.org/developers/
16.4.1 Build report
Report of the build status of your package. It posts at around 11AM EST (can change sometimes) and is located on the time stamp at the top of the page.
Two versions of the build report.
Bioconductor RELEASE version
Bioconductor development version
Six build machines in total at Bioconductor.
DEVEL (3 for devel)
- Linux (malbec1)
- Windows (tokay1)
- OS X (merida1)
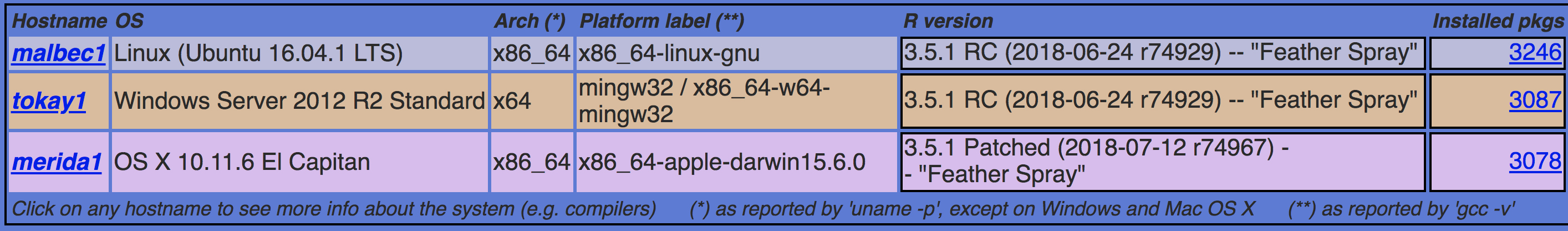
Build machines on DEVEL
RELEASE (3 for release)
- Linux (malbec2)
- Windows (tokay2)
- OS X (merida2)
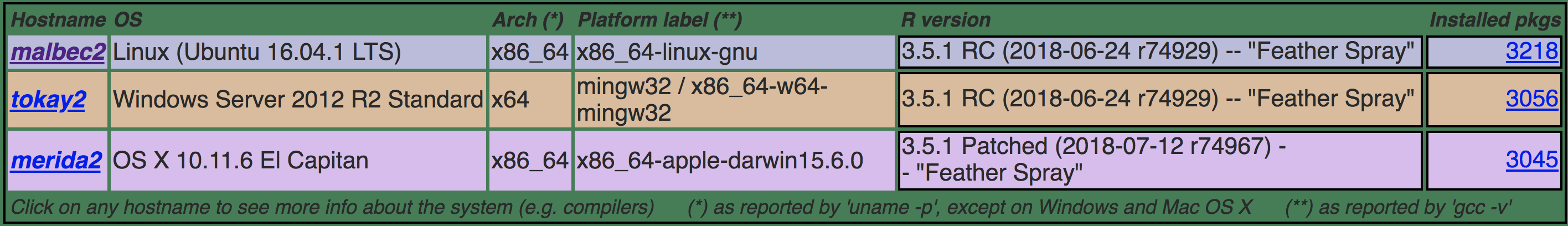
Build machines on RELEASE
16.4.1.1 What is running on these machines?
16.4.1.1.1 INSTALL
Command:
/home/biocbuild/bbs-3.7-bioc/R/bin/R CMD INSTALL a4http://bioconductor.org/checkResults/release/bioc-LATEST/a4/malbec2-install.html
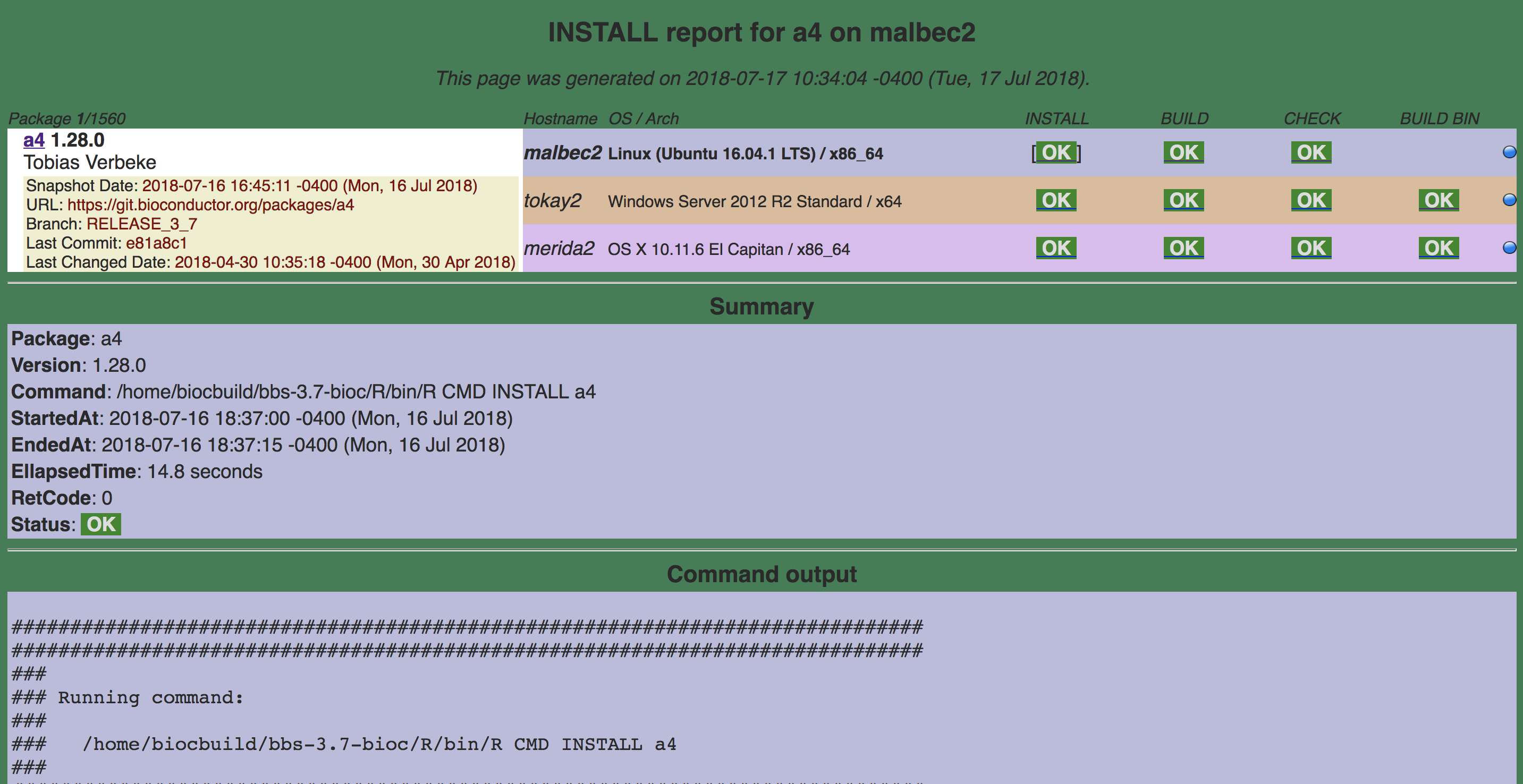
a4 R CMD INSTALL
16.4.1.1.2 BUILD
Command:
/home/biocbuild/bbs-3.7-bioc/R/bin/R CMD build a4http://bioconductor.org/checkResults/release/bioc-LATEST/a4/malbec2-buildsrc.html
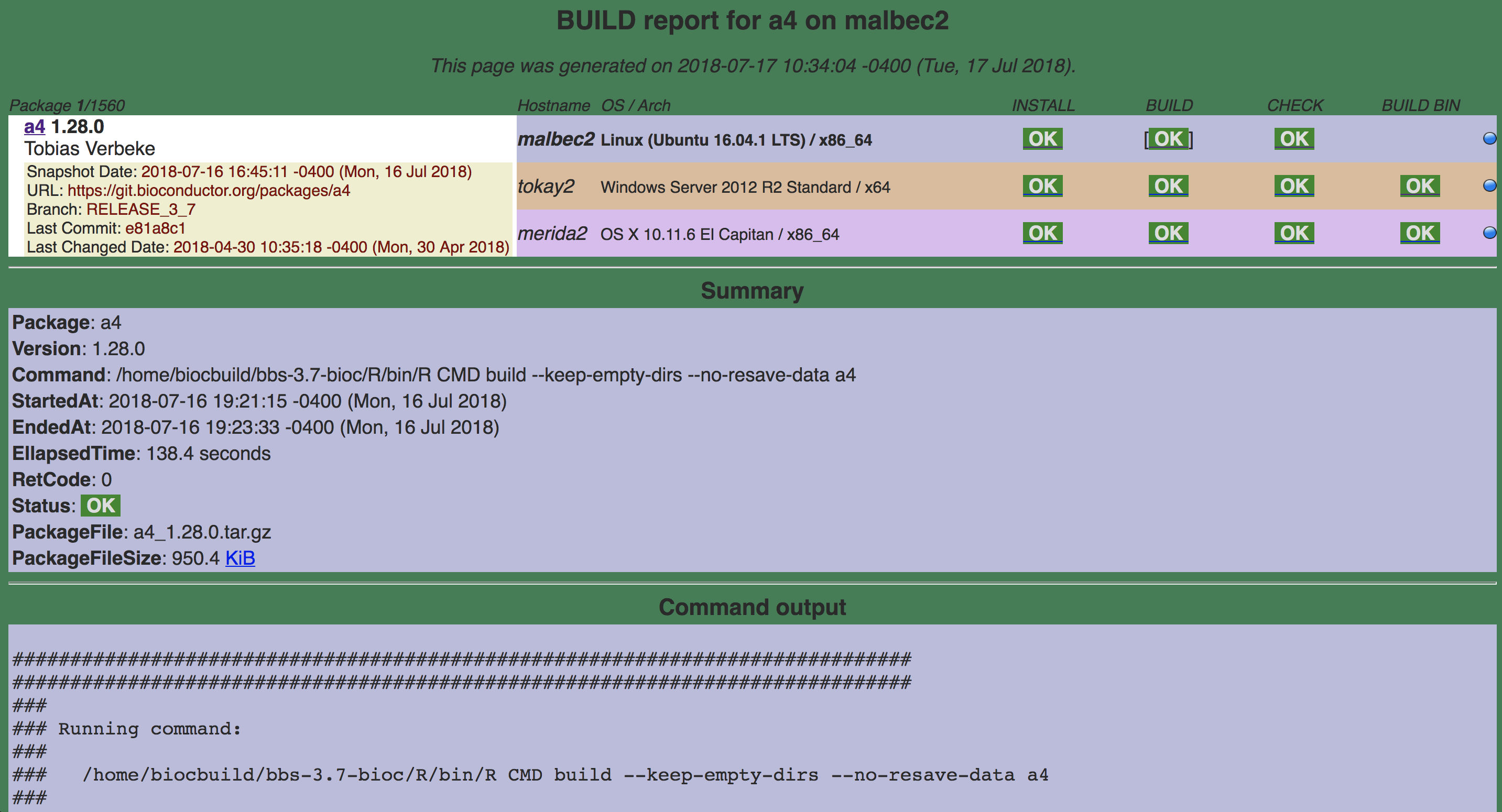
a4 R CMD build
16.4.1.1.3 CHECK
Command:
/home/biocbuild/bbs-3.7-bioc/R/bin/R CMD check a4_1.28.0.tar.gzhttp://bioconductor.org/checkResults/release/bioc-LATEST/a4/malbec2-checksrc.html
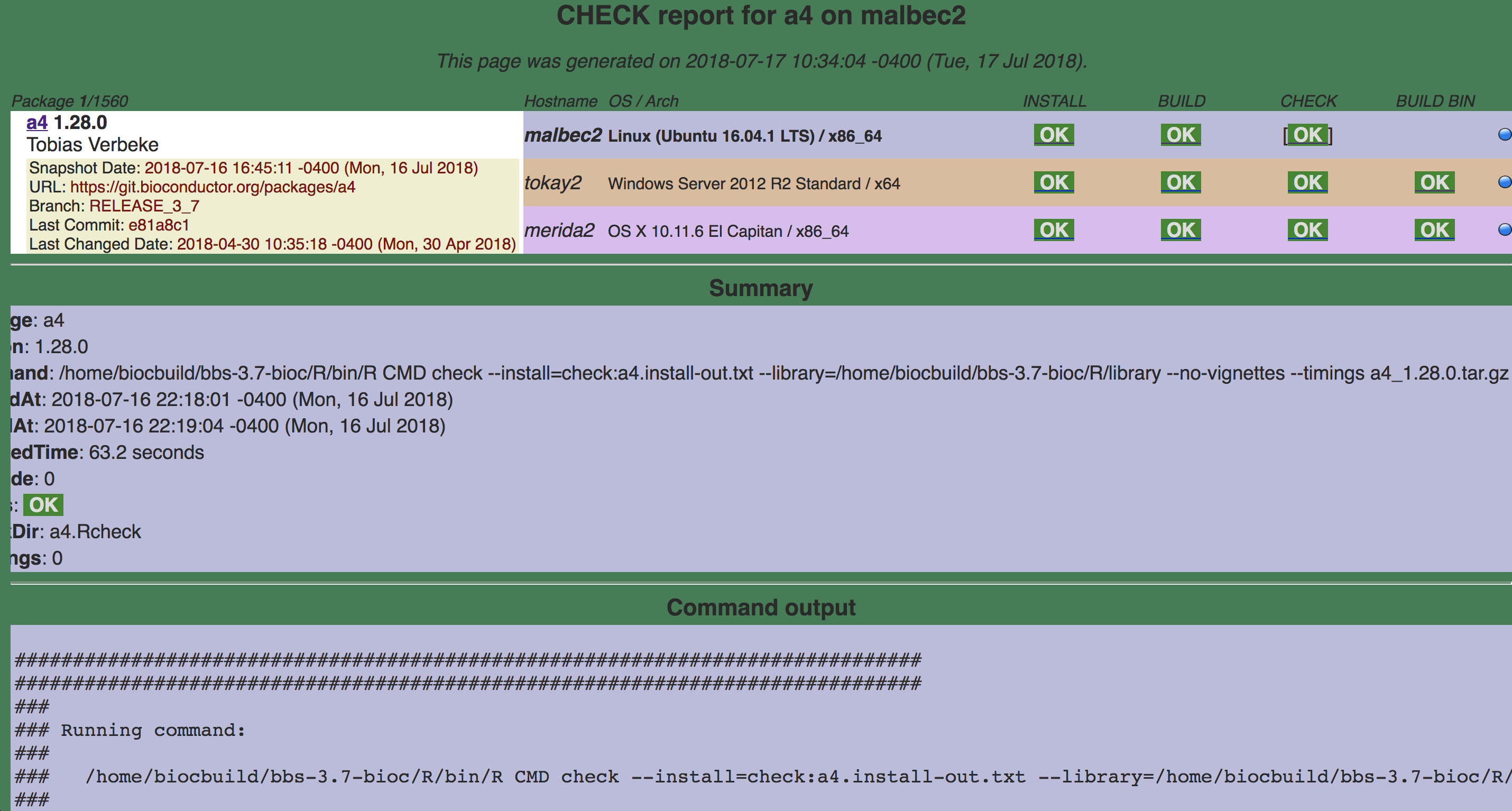
a4 R CMD check
16.4.1.1.4 BUILD BIN
This makes the tar ball which is specific to the platform and propagates it to main repository.
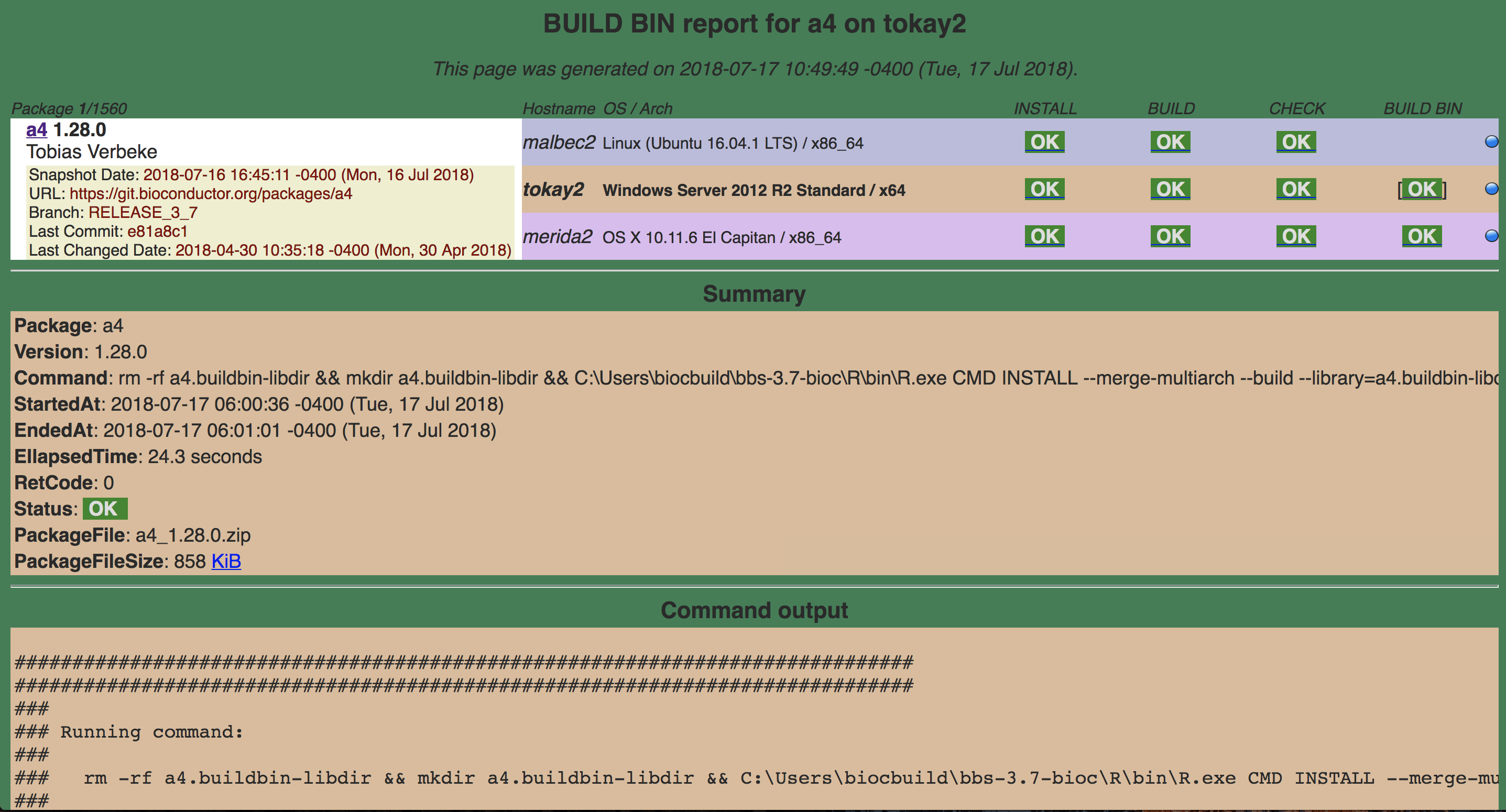
a4 buildbin
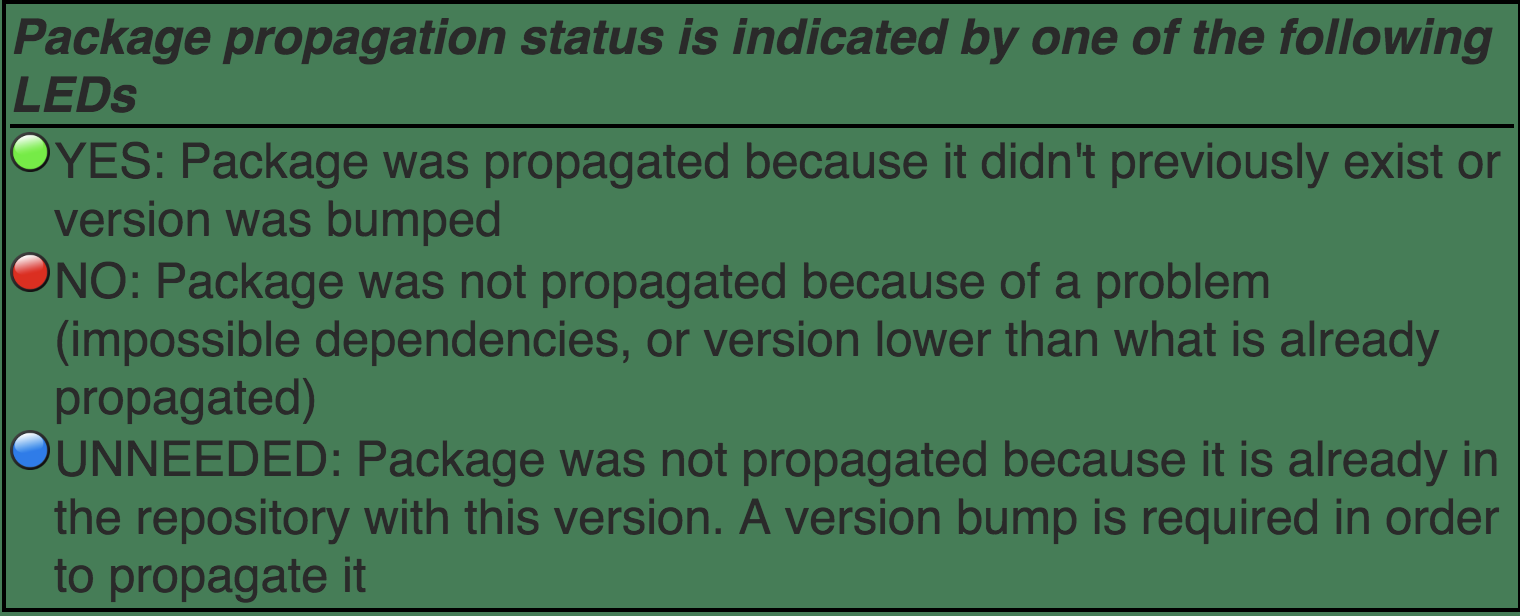
Lights next to the build-bin
The build machines display a status for each package. They indicate different things.
TIMEOUT - INSTALL, BUILD, CHECK or BUILD BIN of package took more than 40 minutes
ERROR - INSTALL, BUILD or BUILD BIN of package failed, or CHECK produced errors
WARNINGS - CHECK of package produced warnings
OK - INSTALL, BUILD, CHECK or BUILD BIN of package was OK
NotNeeded - INSTALL of package was not needed (click on glyph to see why)
skipped - CHECK or BUILD BIN of package was skipped because the BUILD step failed
NA- BUILD, CHECK or BUILD BIN result is not available because of an anomaly in the Build System
NOTE: The build machines run these commands a little differently and the maintainers need not do it the same way.
16.4.1.2 Exercise:
Question:
Filter packages on the DEVEL build machines which have a WARNING on their
build report. List the top 5 packages from this filter.Question:
What is the timestamp on DEVEL build report of the package BiocParallel?16.4.2 Version Numbering
Version numbering is crucial to getting your package to build on the Bioconductor build machines. Every time you want to push a change, it’s important that you make a version number update if you want to your package to propagate to your users. The package is not built unless there is a version number update.
https://bioconductor.org/developers/how-to/version-numbering/
16.4.2.1 Exercise:
Question:
A very common developer question is, "I made a commit and pushed to my
package, but the new build was not triggered. Why wasn't the
build triggered?"
The immediate response is to check if they issued a version bump. How
would someone check this?Question:
What is the version number next to the RELEASE and devel versions
of your package? Do they align with Bioconductor policy as given
on https://bioconductor.org/developers/how-to/version-numbering/ ?16.4.3 Landing page badges
What are badges?
Badges can indicate the HEALTH and realiability of a package. A reason users look at the badges is to get a sense of which packages to use in their analysis. Active maintainers, useful posts and how long the package is in Bioconductor sway users towards packages.
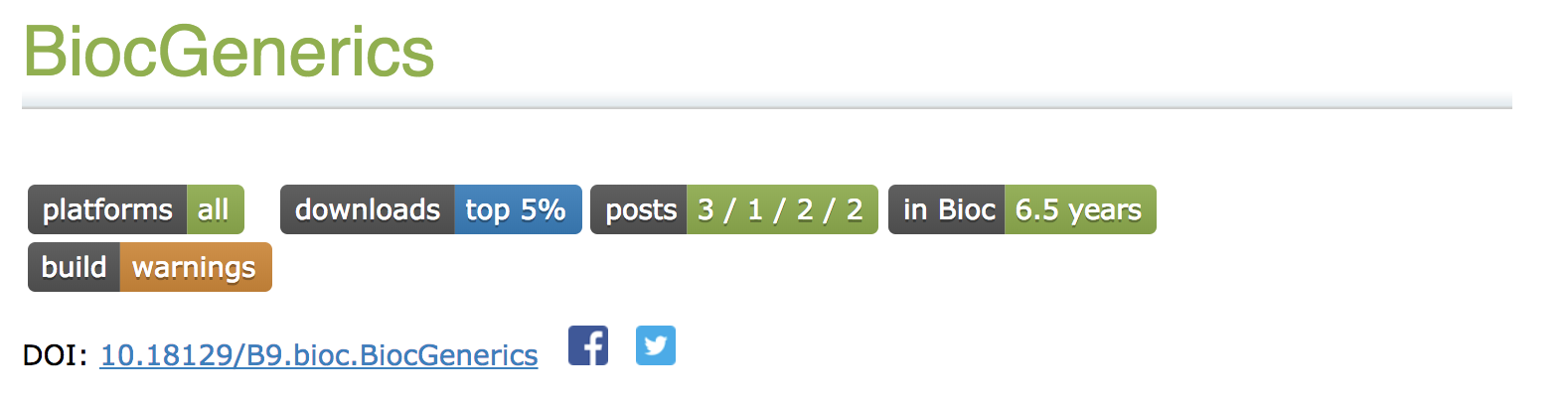
BiocGenerics badges
Badges indicate the following:
platforms availability of the package
downloads statistics
posts on support.bioconductor.org which are related to that package based on the
tag.in Bioc how long the package has been in Bioconductor.
build current build status.
Where can I find these badges?
Landing page to go to
RELEASEbioconductor.org/packages/release/BiocGenerics
Landing page to go to
develbioconductor.org/packages/devel/BiocGenerics
NOTE: After a push is made it can take up to 48 hours for it to appear on the build report and landing pages - i.e. we do one pull, build, check, propagate in 24 hours - so if your commit was after the pull for the day it wont be until the following day
16.4.4 Download statistics
The number reported next to each package name is the download score, that is, the average number of distinct IPs that hit the package each month for the last 12 months (not counting the current month).
http://bioconductor.org/packages/stats/
Top 75 packages as of July 17th 3:15pm EST,

Top 75
BiocGenerics Download statistics,
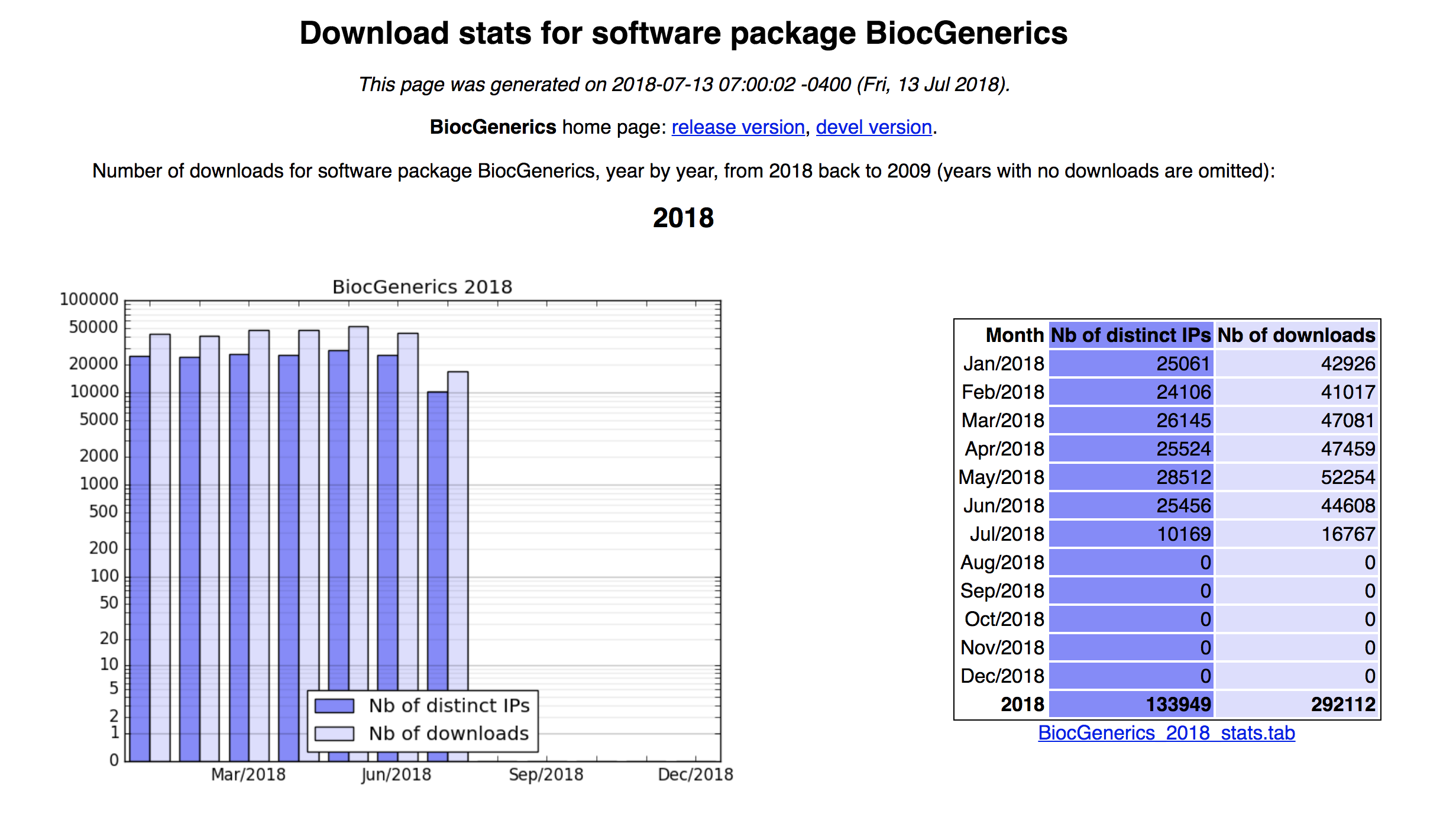
BiocGenerics Download Statistics
16.4.4.1 Exercise:
Question:
Get the download statistics to the package you maintain or the package
you use the most. Next, get the month in 2017 with the most number of
downloads.16.4.5 RSS feeds
RSS feeds help developers keep track of the development going on in Bioconductor across all packages. There are multiple ways to use RSS feeds, and you can use the application of your choice to subscribe to the feeds.
Blog which shows the 12 best RSS reader apps across all platforms.
Links for RSS feeds,
http://bioconductor.org/developers/rss-feeds/
Git log published on the website. The git log hosts the last 500 commits to bioconductor across all packages in the development branch of Bioconductor.
http://bioconductor.org/developers/gitlog/
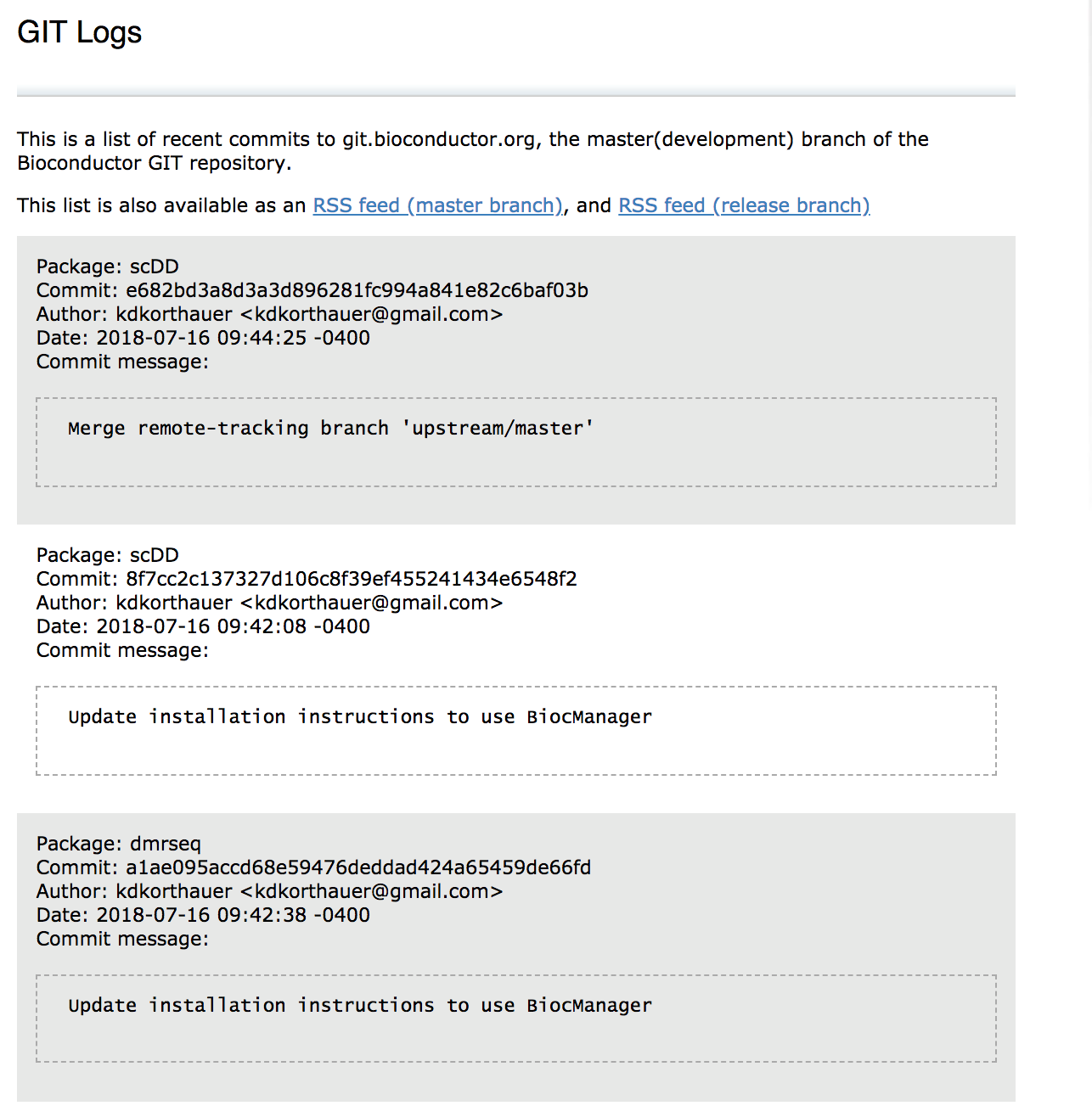
Git log on Jul 16th 2018 at 10:30 AM EST
16.4.5.1 Exercise:
Question:
Take a package of your choice and subscribe to the RSS feed. Use a
tool from the given link, or one of your own liking. (I use
"feedly" the chrome extension)16.4.6 Support site, Bioc-devel mailing list
Support infrastructure set up for Bioconductor users to be in touch with maintainers.
16.4.6.1 Mailing list
The mailing list bioc-devel is for all questions related package development, infrastructure, package support. If a question is misplaced, the author will be pointed to the correct location to ask the question.
16.4.6.2 Support site
Public support site, with easy to use interface to ask questions among the broader community. Any and all end-user questions are welcome here.
16.4.7 GitHub
Optional, but recommended.
16.4.7.1 Sync package with Github / Bioconductor before every change
Do this every time there is an update on your package from your end, or, before you start developing.
Help with Git (most canonical location),
http://bioconductor.org/developers/how-to/git/Sync package from Bioconductor repository to Github repository
http://bioconductor.org/developers/how-to/git/sync-existing-repositories/
16.4.7.2 Github issues
Github issues are more developer centric than mailing list and support site questions. This needs to be posted directly on the development repositories of individual packages. Each package maintainer can choose to do their development on Github giving an interface for issues.
The Bioconductor core team manages issues on the repositories maintained under the Bioconductor Organization on Github.
The packages maintained by Bioconductor core follow the structure
https://github.com/Bioconductor/<PACKAGE_NAME>/issuesExample: https://github.com/Bioconductor/GenomicRanges/issues
16.5 Round up of resources available
Material covered in this workshop very briefly, highlighting
Build machines, what commands the build machines run and what information is displayed on the build reports.
How to check the
HEALTHof your package with badges on the landing pages.Download statistics of packages.
RSS feeds and subscribing to them, checking package development across bioconductor with GIT logs.
Support sites, mailing lists and where to get specific support as a maintainer.
Github and social coding with issues.
There is plenty of infrastructure not covered in this short workshop,
Docker containers.
AWS - Amazon Machine images(AMIs).
BiocCredentials Application for SSH key processing for developers.
Git + Bioconductor private git server.
Single package builder during package contribution.
Github and webhooks for continuous integration.
16.6 Acknowledgements
Roswell Park Comprehensive Cancer Center, Buffalo, NY↩